Orchestrating Self-Contained Applications for Decentralized Data Hubs
Build portable, low-latency data applications that work offline. Transform raw data into fully deployable edge databases with embedded UIs.
What is an Edge Database?
An Edge Database is a database designed to operate at the network "edge"—meaning close to the user, the source of data generation (like a research lab, IoT device, or a local machine). Unlike traditional cloud databases, the primary focus is on low-latency access and offline capability. They prioritize portability and execution speed over centralized control.
Traditional Cloud Databases
- • High latency due to necessary network round trips
- • Requires persistent, reliable internet connection
- • Scales by provisioning larger remote data centers (vertical scaling)
Edge Databases
- • Extremely low latency (data is local to the application)
- • Works entirely offline or with intermittent connectivity
- • Scales by distributing self-contained, identical copies
- • Uses portable formats like SQLite
Case Study: Diabetes Research Hub (DRH) Edge Data Pipeline
In the Diabetes Research Hub (DRH) project, the final artifact, a single resource-surveillance.sqlite.db file, is the Edge Database. It contains both the processed research data and the entire web application UI (powered by SQLPage) necessary to view it, making it a fully portable, "database-as-an-app" solution suitable for clinic and research settings.
The Spry framework orchestrates the complete ETL (Extract, Transform, Load) and deployment pipeline, defined in the drh-simplera-spry.md manifest. It manages the execution of specialized tools to convert raw study files (CGM, meal, fitness data) into the final Edge Database.
Core Tools and Technologies
Spry
Task Orchestration & Dependency Management, the 'glue' of the pipeline.
surveilr
Initial data ingestion, format conversion (CSV to RSSD), and pre-processing.
Spry Based WorkBook Execution
Individual Task Execution
spry rb task [taskname] [markdownfilename]
Run a specific task defined in the markdown file independently.
Execute Full WorkBook
spry rb run [markdownfilename]
All tasks in the markdown can be executed in one go.
The Multi-task Pipeline Breakdown
Task: prepare-env
Role : Initializes the execution environment by checking for mandatory tools and ensuring all required environment variables are sourced (like SPRY_DB, STUDY_DATA_PATH, TENANT_ID).
```bash prepare-env -C ./.envrc --gitignore --descr "Generate .envrc file and add it to local .gitignore"
export SPRY_DB="sqlite://resource-surveillance.sqlite.db?mode=rwc"
export PORT=9227
export STUDY_DATA_PATH="raw-data/simplera-synthetic-cgm/"
export TENANT_ID="FLCG"
export TENANT_NAME="Florida Clinical Group"
direnv allow
```Task: prepare-db-deploy-server
Automated Pipeline: Performs pre-etl-validation, ingestion, complex DuckDB ETL (conditional on validation status), and initializes the SQLPage server.
```bash
#!/bin/bash
set -u
# 1. Cleanup
rm -f resource-surveillance.sqlite.db *.sql
rm -rf dev-src.auto validation-reports
# 2. RUN PREFLIGHT VALIDATION
surveilr ingest files -r "${STUDY_DATA_PATH}" --tenant-id "${TENANT_ID}" --tenant-name "${TENANT_NAME}"
surveilr shell --engine duckdb duckdb-etl-sql/drh-preflight-validation.sql
# 3. EXTRACT STATUS FROM JSON
RAW_STATUS=$(surveilr shell "select overall_status from drh_validation_reports ORDER BY timestamp DESC LIMIT 1;")
VALIDATION_STATUS=$(echo "$RAW_STATUS" | jq -r '.[0].overall_status')
# 4. CONDITIONAL ETL EXECUTION
if [ "$VALIDATION_STATUS" == "PASS" ]; then
(
set -e
surveilr orchestrate transform-csv
surveilr shell common-sql/drh-data-validation.sql
surveilr shell common-sql/drh-anonymize-prepare.sql
surveilr shell --engine duckdb duckdb-etl-sql/drh-master-etl.sql
surveilr shell common-sql/drh-metrics-pipeline.sql
)
[ $? -ne 0 ] && exit 1
else
echo "Validation FAILED ($VALIDATION_STATUS). Skipping ETL steps."
fi
# 5. INITIALIZE SQLPAGE
spry sp spc --package --conf sqlpage/sqlpage.json -m drh-dexcom-cgm-spry.md | sqlite3 resource-surveillance.sqlite.db
```Run the server
Role: The final deployment. This executes the pre-compiled SQLPage binary, pointing it directly to the fully compiled SQLite database. The server instantly reads the embedded UI and data, launching the complete, interactive DRH application.
Execution Command: sqlpage
```bash
sqlpage
```Why This Approach Works
- • Single-File Portability: The entire application—data, ETL logic, and UI—exists in one SQLite file that can be copied to any machine.
- • Zero-Latency Access: All queries execute locally with sub-millisecond response times, perfect for clinical environments with unreliable connectivity.
- • Reproducible Pipelines: The entire transformation process is codified in version-controlled Markdown, ensuring consistent results across research sites.
- • Embedded UI: SQLPage transforms the database into a web server, eliminating the need for separate deployment infrastructure.
Application Screenshots (DRH UI)
The following images illustrate the final interactive web application compiled and served by the SQLPage binary using the embedded Edge Database.
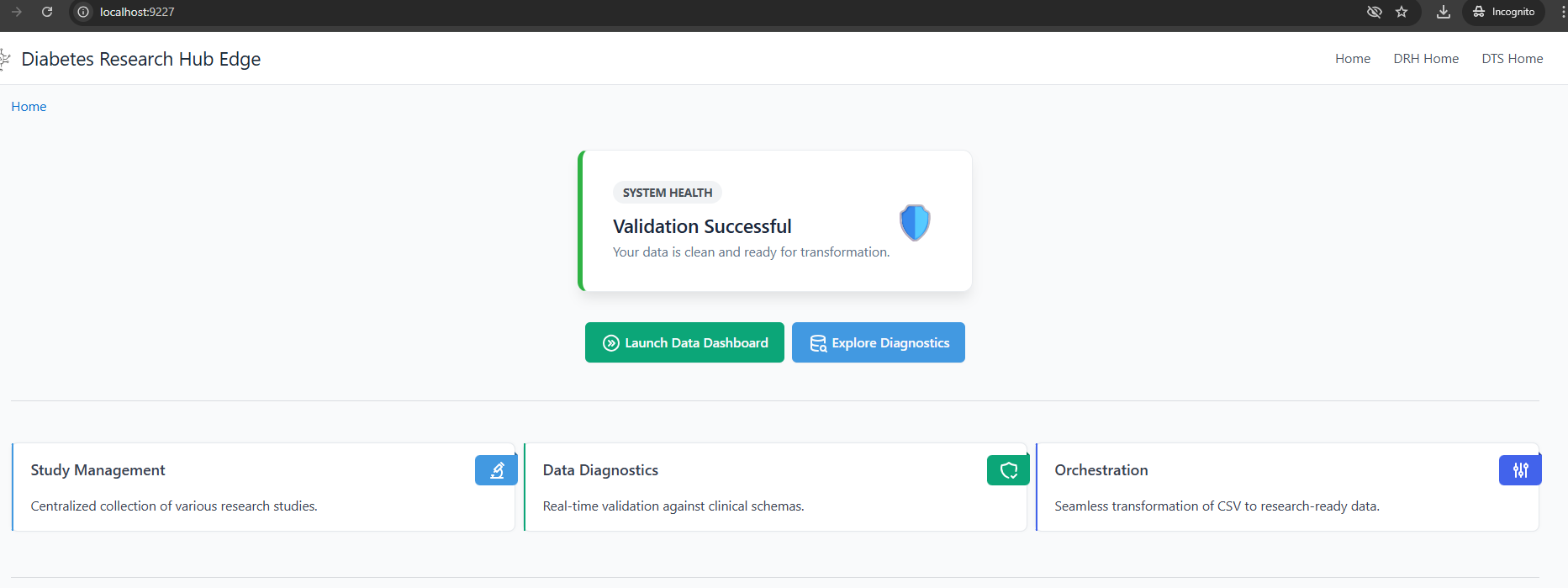
DRH Home
Note: The user interface is dynamically generated by SQLPage, querying the data tables created in the prepare-db stage. [cite: 69]